Probability of Detection (Pdet) generation and comparison
This notebook is a full guide on how to use the gwsnr package to generate the Pdet for a given Gravitational Wave (GW) signal parameters.
Contents of this notebook
Calculation by considering \(P_{\rm det}\) as, - Boolean (detected if 1, not detected if 0) - with \(\rho_{\rm obs}\) as non-central \(\chi\) distribution - with \(\rho_{\rm obs}\) as Gaussian distribution - with \(\rho_{\rm opt}\) as Fixed SNR. - Probabilistic (between 0 and 1) - with \(\rho_{\rm obs}\) as non-central \(\chi\) distribution - with \(\rho_{\rm obs}\) as Gaussian distribution
For snr generation, in this example notebook, I will use the default snr_method, i.e. ‘interpolation_aligned_spins’. Please refer to the snr_generation.ipynb for more details on snr generation methods.
‘pdet_kwargs’ can be initialized while creating the GWSNR class object or the pdet related parameters can be passed while calling the pdet calculation method.
Note: For more details on Pdet calculation methods, please refer to the gwsnr documentation.
[98]:
import numpy as np
import matplotlib.pyplot as plt
from datetime import datetime
import gwsnr
np.random.seed(42)
gwsnr = gwsnr.GWSNR(
npool=4,
snr_method='interpolation_aligned_spins',
gwsnr_verbose=False,
waveform_approximant='IMRPhenomD',
# pdet_kwargs=dict(snr_th=10.0, snr_th_net=10.0, pdet_type='boolean', distribution_type='noncentral_chi2') # 'pdet_kwargs' can be initialized while creating the GWSNR class object or the pdet related settings can be passed while calling the pdet calculation method.
)
mass_1 = np.array([2,50.,100.,])
ratio = 0.9
dl = 500
# pdet with default settings
pdet = gwsnr.pdet(mass_1=mass_1, mass_2=mass_1*ratio, luminosity_distance=dl)
print("Network Pdet (boolean, non-central chi2) : ", pdet['pdet_net'])
Initializing GWSNR class...
psds not given. Choosing bilby's default psds
Interpolator will be loaded for L1 detector from ./interpolator_pickle/L1/partialSNR_dict_3.pickle
Interpolator will be loaded for H1 detector from ./interpolator_pickle/H1/partialSNR_dict_3.pickle
Interpolator will be loaded for V1 detector from ./interpolator_pickle/V1/partialSNR_dict_3.pickle
Network Pdet (boolean, non-central chi2) : [0 1 1]
[102]:
# get `ler` package generated GW parameters for BBH
!wget https://raw.githubusercontent.com/hemantaph/gwsnr/refs/heads/main/tests/integration/bbh_gw_params.json
--2025-10-28 13:12:44-- https://raw.githubusercontent.com/hemantaph/gwsnr/refs/heads/main/tests/integration/bbh_gw_params.json
Resolving raw.githubusercontent.com (raw.githubusercontent.com)... 185.199.110.133, 185.199.109.133, 185.199.108.133, ...
Connecting to raw.githubusercontent.com (raw.githubusercontent.com)|185.199.110.133|:443... connected.
HTTP request sent, awaiting response... 200 OK
Length: 4986213 (4.8M) [text/plain]
Saving to: ‘bbh_gw_params.json’
bbh_gw_params.json 100%[===================>] 4.75M 16.5MB/s in 0.3s
2025-10-28 13:12:44 (16.5 MB/s) - ‘bbh_gw_params.json’ saved [4986213/4986213]
[103]:
# # delete the downloaded json file
# !rm bbh_gw_params.json
[104]:
from gwsnr.utils import get_param_from_json
param_dict = get_param_from_json('bbh_gw_params.json')
Boolean Pdet calculation
Non-Central \(\chi\) distribution
[105]:
pdet_chi2 = gwsnr.pdet(
gw_param_dict=param_dict,
snr_th_net=10.0,
pdet_type='boolean',
distribution_type='noncentral_chi2',
include_observed_snr=True, # to include observed SNR values in the output dictionary
)
# print the first 5 pdet values
print(pdet_chi2['pdet_net'][:5])
print(pdet_chi2['observed_snr_net'][:5])
# find and print detectable fraction
detectable_fraction = np.mean(pdet_chi2['pdet_net'])
print("Detectable fraction of the population : ", detectable_fraction)
[0 0 1 0 0]
[ 3.16872665 3.63206794 13.38517257 2.50378508 2.62985054]
Detectable fraction of the population : 0.0036
Gaussian distribution
[20]:
pdet_gauss = gwsnr.pdet(
gw_param_dict=param_dict,
snr_th_net=10.0,
pdet_type='boolean',
distribution_type='gaussian',
include_observed_snr=True
)
# print the first 5 pdet values
print(pdet_gauss['pdet_net'][:5])
print(pdet_gauss['observed_snr_net'][:5])
# find and print detectable fraction
detectable_fraction = np.mean(pdet_gauss['pdet_net'])
print("Detectable fraction of the population : ", detectable_fraction)
[0 1 1 0 0]
[ 4.87782373 15.50141248 14.35847191 3.64935961 2.98302797]
Detectable fraction of the population : 0.34
Fixed SNR
[21]:
pdet_optsnr = gwsnr.pdet(
gw_param_dict=param_dict,
snr_th_net=10.0,
pdet_type='boolean',
distribution_type='fixed_snr',
include_observed_snr=True, # here observed_snr_net will be same as optimal SNR
)
# print the first 5 pdet values
print(pdet_optsnr['pdet_net'][:5])
print(pdet_optsnr['observed_snr_net'][:5])
# find and print detectable fraction
detectable_fraction = np.mean(pdet_optsnr['pdet_net'])
print("Detectable fraction of the population : ", detectable_fraction)
[0 1 1 0 0]
[ 4.21494246 14.32793862 14.17745035 4.94619156 2.58334002]
Detectable fraction of the population : 0.32
Visulalization and Comparison Observed SNR distributions wrt to Optimal SNR
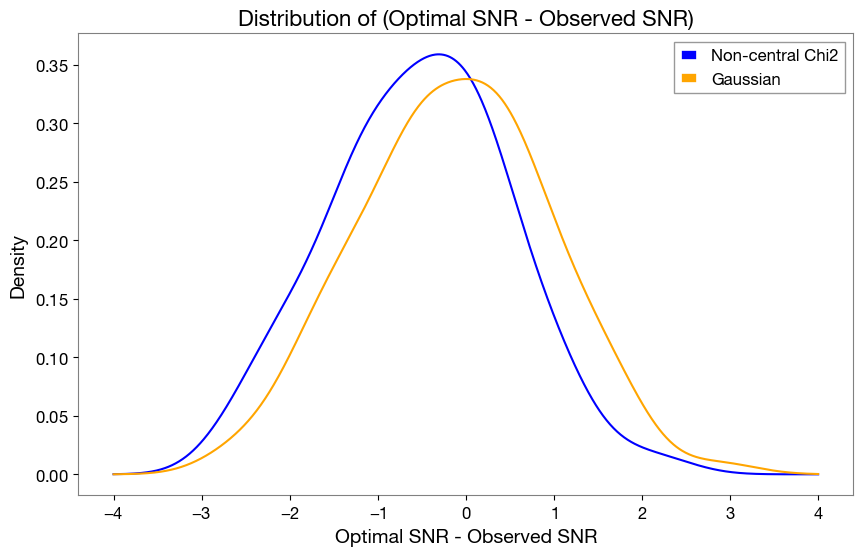
[25]:
snr_opt = pdet_optsnr['observed_snr_net']
snr_obs_chi2 = pdet_chi2['observed_snr_net']
snr_obs_gauss = pdet_gauss['observed_snr_net']
dist1 = snr_opt - snr_obs_chi2
dist2 = snr_opt - snr_obs_gauss
# create kde plots to compare the two distributions
from scipy.stats import gaussian_kde
kde1 = gaussian_kde(dist1)
kde2 = gaussian_kde(dist2)
x = np.linspace(-4, 4, 1000)
plt.figure(figsize=(10,6))
# plt.hist(dist1, bins=30, alpha=0.5, label='Non-central Chi2', color='blue', density=True, histtype='step')
# plt.hist(dist2, bins=30, alpha=0.5, label='Gaussian', color='orange', density=True, histtype='step')
plt.plot(x, kde1(x), label='Non-central Chi2', color='blue')
plt.plot(x, kde2(x), label='Gaussian', color='orange')
plt.title('Distribution of (Optimal SNR - Observed SNR)', fontsize=16)
plt.xlabel('Optimal SNR - Observed SNR', fontsize=14)
plt.ylabel('Density', fontsize=14)
plt.legend(fontsize=12)
plt.grid()
plt.show()
Probabilistic Pdet calculation
Non-Central \(\chi\) distribution
[27]:
pdet_chi2 = gwsnr.pdet(
gw_param_dict=param_dict,
snr_th_net=10.0,
pdet_type='probability_distribution',
distribution_type='noncentral_chi2',
include_optimal_snr=True, # to include optimal SNR values in the output dictionary
)
# print the first 5 pdet values
print(pdet_chi2['pdet_net'][:5])
print(pdet_chi2['optimal_snr_net'][:5])
# find and print detectable fraction
detectable_fraction = np.mean(pdet_chi2['pdet_net'])
print("Detectable fraction of the population : ", detectable_fraction)
[3.13236730e-08 9.99997133e-01 9.99994244e-01 1.26963490e-06
1.70041758e-12]
[ 4.21494246 14.32793862 14.17745035 4.94619156 2.58334002]
Detectable fraction of the population : 0.3272490373160273
Gaussian distribution
[28]:
pdet_gauss = gwsnr.pdet(
gw_param_dict=param_dict,
snr_th_net=10.0,
pdet_type='probability_distribution',
distribution_type='gaussian',
include_optimal_snr=True
)
# print the first 5 pdet values
print(pdet_gauss['pdet_net'][:5])
print(pdet_gauss['optimal_snr_net'][:5])
# find and print detectable fraction
detectable_fraction = np.mean(pdet_gauss['pdet_net'])
print("Detectable fraction of the population : ", detectable_fraction)
[3.62437580e-09 9.99992474e-01 9.99985260e-01 2.16542995e-07
6.00630656e-14]
[ 4.21494246 14.32793862 14.17745035 4.94619156 2.58334002]
Detectable fraction of the population : 0.32040024945890544
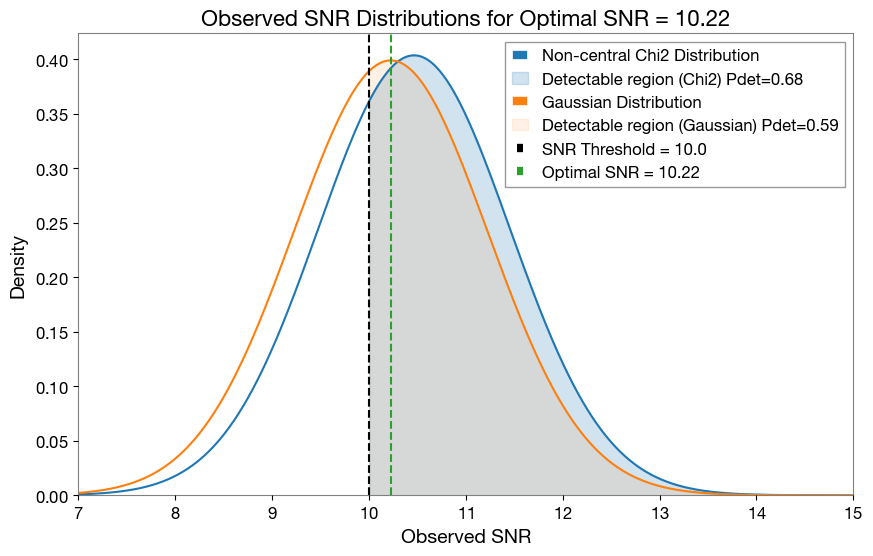
[97]:
### Visulalization Pdet based on Probability distribution of Observed SNR wrt to Optimal SNR
from scipy.stats import norm, ncx2
min_snr = 7
max_snr = 15
x_values = np.linspace(min_snr, max_snr, 1000)
# pdet_gauss['pdet_net'] between 0.2 and 0.5
selected_indices = np.where((pdet_gauss['pdet_net'] >= 0.5) & (pdet_gauss['pdet_net'] <= 0.9))[0]
selected_index = selected_indices[0]
# Non-central Chi2 distribution case
size = 1000
nc_param = pdet_chi2['optimal_snr_net'][selected_index]**2
df = 2 * len(gwsnr.detector_list)
pdf_dist1 = ncx2.pdf(x_values**2, df=df, nc=nc_param)
print(np.trapz(pdf_dist1, x_values**2))
# normalize the pdf; or you might need to take care of Jacobian while plotting
# pdf_dist1 /= np.trapz(pdf_dist1, x_values)
# Gaussian distribution case
size = 1000
mu = pdet_gauss['optimal_snr_net'][selected_index]
sigma = 1.0 # standard deviation
pdf_dist2 = norm(loc=mu, scale=sigma)
pdf_dist2 = pdf_dist2.pdf(x_values)
print(np.trapz(pdf_dist2, x_values))
# pdf_dist2 /= np.trapz(pdf_dist2, x_values)
plt.figure(figsize=(10,6))
plt.plot(x_values, pdf_dist1 * 2 * x_values, label='Non-central Chi2 Distribution', color='C0')
plt.fill_between(x_values, 0, pdf_dist1 * 2 * x_values, where=(x_values >= 10.0), alpha=0.2, color='C0', label=f'Detectable region (Chi2) Pdet={pdet_chi2["pdet_net"][selected_index]:.2f}')
plt.plot(x_values, pdf_dist2, label='Gaussian Distribution', color='C1')
plt.fill_between(x_values, 0, pdf_dist2, where=(x_values >= 10.0), alpha=0.1, color='C1', label=f'Detectable region (Gaussian) Pdet={pdet_gauss["pdet_net"][selected_index]:.2f}')
plt.title('Observed SNR Distributions for Optimal SNR = {:.2f}'.format(pdet_gauss['optimal_snr_net'][selected_index]), fontsize=16)
# vertical line for SNR threshold
plt.axvline(x=10.0, color='k', linestyle='--', label='SNR Threshold = 10.0')
plt.axvline(x=pdet_chi2['optimal_snr_net'][selected_index], color='C2', linestyle='--', label=f'Optimal SNR = {pdet_chi2["optimal_snr_net"][selected_index]:.2f}')
plt.legend(fontsize=12, loc='upper right')
plt.xlim(7, 15)
plt.ylim(0, None)
plt.xlabel('Observed SNR', fontsize=14)
plt.ylabel('Density', fontsize=14)
plt.grid()
plt.show()
0.9997810824868587
0.9993681083703562
[100]:
# delete the downloaded json file
!rm bbh_gw_params.json